Patient-Specific Survival Prediction (PSSP)
The most commonly used approach for predicting survival times is to create a survival curve for each category of patient (often using the Kaplan-Meier estimator) such as the curve shown in Figure 1.
The problem with this approach is that it aggregates individual patient characteristics. Our system provides truly personalized predictions of patient survival times. Figures 2 and 3 show PSSP produced predicted survival times for 2 patients who would receive the same predicted survival time using an aggregated approach (as they were both stage 4 stomach cancer). However, when examined individually, we see that PSSP (accurately) predicted these patients to have very different survival curves. When looking at many patient survival curves at once (Figure 4), we can see that an aggregate approach would obscure the wide range of patient-specific survival curves.
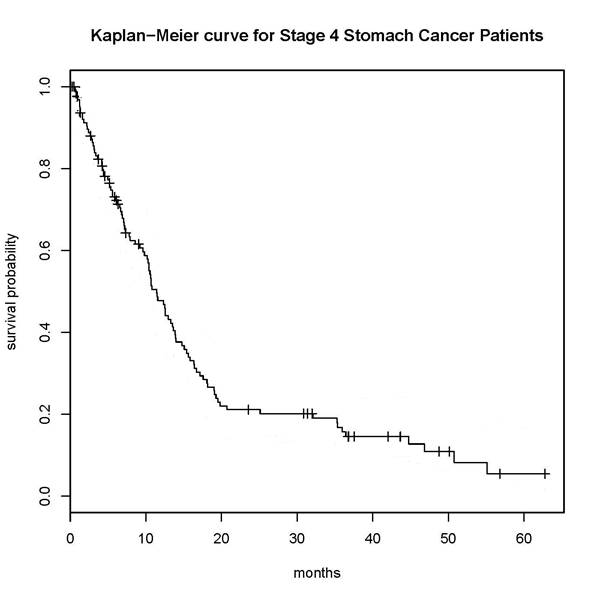 Fig. 1 |
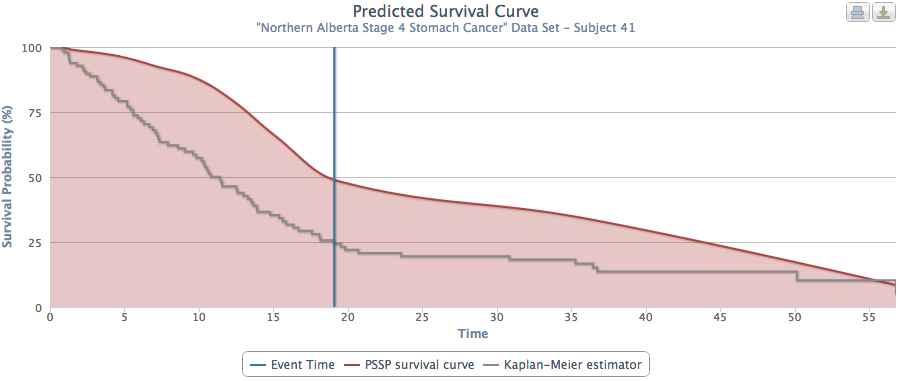 Fig. 2 |
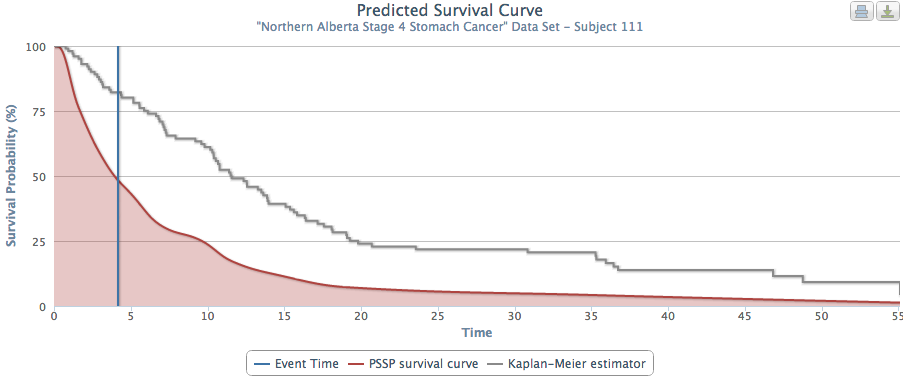 Fig. 3 |
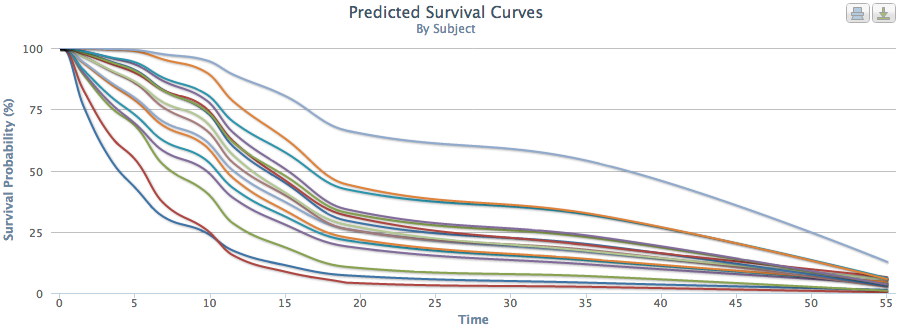 Fig. 4 |
||
This website provides access to the PSSP codebase. You can either analyze your data set on this site, or download the source for the command-line PSSP tool. To better understand PSSP, see the slides and presentation here; and how to use this website, see the Tutorial.
You can also look at our publicly accessible predictors here.
Learning Patient-Specific Survival Distributions
as a Sequence of Dependent Regressors
Chun-Nam Yu (Postdoctoral Fellow),
Russ Greiner (Professor),
Vickie Baracos (Professor),
An accurate model of patient survival time can help in the treatment and care of patients. The common practice of providing survival time estimates based only on population averages for the site and stage of the disease ignores many important individual differences among patients. Here, we present a novel machine learning algorithm, PSSP (for “patient-specific survival predictor"), for learning patient-specific survival time distribution based on patient attributes, such as blood tests and clinical assessments. The predicted distribution can be regarded as a personalized version of Kaplan-Meier curve, and can be used as a tool for doctors to visualize the survival rate of individual patients. PSSP can also easily incorporate the time-varying effects of prognostic factors and handle censored survival times. When tested on a cohort of more than 2000 cancer patients from northern Alberta, our method gives survival time predictions that are much more accurate than popular survival analysis models such as the Cox and Aalen regression models. Our results show that using patient-specific attributes can reduce the prediction error on survival time by as much as 20% when compared to using cancer site and stage only. We anticipate this same technology can be used for learning and predicting personalized Kaplan-Meier curves for patients suffering from other diseases.


